|
|
1.IntroductionThe New Zealand Land Cover Database1 (LCDB) is a digital thematic map of land cover and land use. It is mainly produced by manual digitization using satellite imagery and aerial photographs. In the large and relatively homogenous pasture areas, small patches of woody vegetation are often not identified correctly by human interpretation. An automated, or at least a semiautomated, approach for woody/nonwoody classification using imagery that is available nation-wide would, therefore, be desirable in order to improve map quality. As the major aim of this study is to improve the information content of an existing dataset, such a method must be “conservative” in the sense that it only adds new polygons that are woody with a high probability. The improvement of manually digitized land cover maps is a problem that is not specific to the LCDB and the method could, therefore, also be applied to regional and national land cover maps in other regions of the world. Current methods for automated land cover mapping on the regional to national scale are well established for single-source approaches using synthetic aperture radar (SAR) and optical datasets2 as well as synergistic use of SAR and optical imagery that generally produces improved classification results.3–7 A wide range of supervised classification methods are described in this literature. These include statistical methods, such as maximum likelihood and discriminant analysis, machine-learning methods, such as decision tree classifiers, neural networks, and support vector machines8 (SVMs). A number of studies point out that SVMs are effective in classifying high-dimensional input datasets.9–12 Mountrakis et al.13 even conclude that “past applications of the method on both real-world data and simulated environments have shown that SVMs exhibit superiority over most of the alternative algorithms.” Geographic object-based image analysis provides advantages over pixel-based techniques14–17 and is well suited to analyses that combine different data sources as image registration differences average out over segments. The use of light detection and ranging (LiDAR)-derived canopy height models (CHMs) for forest mapping is equally well established with the promise of high spatial resolution and associated high accuracy. Classification results can be improved by combination with data from other sensors.2 However, LiDAR data are rarely available as full coverage for regional and national mapping initiatives due to the high cost. Nevertheless, for small-scale mapping, it is valuable as a training data, at least if the features to be mapped can be well identified by the LiDAR-derived products. It is surprising that despite the existence of methods for automated land cover mapping on regional to national scales, many mapping programs are still mainly created using manual digitization techniques—e.g., the LCDB of New Zealand and the “Peta Penutupan Lahan”18 of Indonesia. The two main reasons for this are (1) the general belief that manual mapping using aerial photography is able to provide better spatial accuracy than automated classification of satellite images and (2) the need for continuity of multitemporal maps. Therefore, approaches are needed which enable automated classification to improve map quality without replacing the manual methods completely. Improvement of map quality in this context refers to the addition of information content from automated methods to the manually derived maps. This approach can also be applied to mapping programs that consider introducing new data sources, e.g., if SAR data are added to a mapping approach that is purely based on optical imagery. In this paper, we describe a segmentation-based method for automated detection and classification of so far unmapped woody patches with a minimum mapping size of 1 ha. A LiDAR CHM is used to train the classifier regarding the woody/nonwoody discrimination, while the classification of identified woody patches uses the existing LCDB (map) in the training step. The classifier makes synergetic use of Phased Array type L-band SAR (PALSAR) and Satellite Pour l’Observation de la Terre (SPOT) data in both training steps. We describe which of the data sources are most able to differentiate between woody and nonwoody vegetation. 2.Data2.1.Land Cover DatabaseThe New Zealand LCDB is a multitemporal digital thematic map of land cover and land use. Land cover at each of four periods—summer 1996/1997, summer 2001/2002, summer 2008/2009, and summer 2012/2013—is both delineated and classified using SPOT imagery and aerial photography. The land cover classification evolved over the first three periods, but compatibility has been maintained. LCDB1 polygons were largely drawn manually by visual interpretation, while later, change polygons were either automatically created or manually drawn. The current version, LCDB v4 (and its predecessor LCDB v3), contains 33 classes. The minimum mapping unit is 1 ha. 2.2.SPOT-5SPOT-5 scenes were acquired during the summers of 2008/2009 and 2012/2013 that covered the whole of New Zealand. All individual scenes were corrected for the effects of atmosphere and illumination19 and then mosaicked together in a prioritized sequence to maximize the image quality while minimizing the amount of viewable cloud. A further standardized reflectance product was then produced in a similar manner where an additional topographic correction stage was performed.19 Both the cloud minimization and the standardized reflectance mosaics were accompanied by a “control” mask that defined which portions of the individual satellite scenes were used in the appropriate mosaic. The green band was not included in the analysis as it generally shows strong correlation with the red band.20 2.3.PALSAR Global MosaicThe PALSAR Global Mosaic21 is a high-resolution slope-corrected and ortho-rectified L-band gamma-naught map. The data used for this study were acquired at 10-m resolution in a fine-beam dual-polarization mode (HH and HV) for the year 2007. The data were resampled to the same grid and coordinate reference system (New Zealand Transverse Mercator; NZTM) as the SPOT-5 imagery. They were then mosaicked and calibrated to power values using calibration factors from the study of Shimada et al.22 and the following equation: where DN is the 16-bit unsigned value from the ALOS-PALSAR product.A conversion to decibels (dB) was not applied at this stage as it was intended to calculate the mean of the backscatter values for later defined segments; therefore, a logarithmic scale would be inappropriate. The data were then reprojected to NZTM and geometrically corrected using a first-order polynomial and 250 GCPs to match the SPOT-5 imagery. 2.4.LiDAR Canopy Height ModelThe LiDAR data used in this study were acquired over the entire Wellington region extending to 100-m offshore, an area slightly larger than 8500 km2. The majority of the LiDAR survey was performed in early 2013 with some additional aircraft flights later in 2013 and 2014, depending on weather and data quality considerations. The LiDAR scanner was an Optech ALTM 3100EA flown at a nominal height of 1000 m. The target survey point density was 1.73 ppsm with 50% swath overlap to ensure the minimum raw data specification of 1.3 ppsm and vertical accuracy of (1 sigma). The 1261 LiDAR flight lines of raw point cloud data were merged, tiled, and automatically classified into ground and nonground returns using the open-source LiDAR processing software, Sorted Pulse Data Software Library.23 The tiles of ground classified points were then interpolated and mosaicked into a 1-m resolution digital terrain model (DTM). Similarly, the tiles of nonground classified points were also interpolated and mosaicked at 1-m resolution from which the DTM is subtracted to form a CHM. The term CHM is nominal as it is primarily derived from canopy returns but will contain other identified nonground features such as buildings in urban areas. 3.MethodsWe used a two-step classification with a first step differentiating between woody and nonwoody and a second step classifying identified woody patches according to the existing LCDB classes. 3.1.Identification of Woody PatchesWe first segmented the SPOT imagery using an algorithm called the iterative elimination method available in the Remote Sensing and GIS Software Library.24 The algorithm uses K-means clustering25 for seeding and classifying pixels in a first step. Then the classified pixels are clumped into geographically uniquely labeled regions and small clumps are merged iteratively with their most similar larger neighboring segments until the minimum mapping size is reached. Segmentation was based on the red, near-infrared (NIR), and short-wave infrared (SWIR) band of both SPOT mosaics and the minimal segment size was set to 1 ha, according to the minimum mapping unit, which resulted in 277,838 segments in the Wellington region. To allow further analysis, the segments were then associated with statistics relevant to CHM, SPOT, and PALSAR imagery as well as a prerelease of New Zealand LCDB with the version 4.1. The calculated statistics are as follows:
To derive training data, we used the CHM to identify the segments that are clearly woody (more than 90% covered by CHM values larger than 3 m; 58,063 segments) and those that are clearly nonwoody (less than 10% covered by CHM values larger than 0.5 m; 82,236 segments). The thresholds for defining woody areas were chosen for the following reasons. First, we wanted to have homogenous sets of segments as a training dataset; vegetation that is higher than 3 m is certainly woody in the New Zealand context and confusion with agricultural plants such as maize and vineyards as well as flax can be avoided. The second reason for the 90% threshold is that the CHM includes built-up areas. Segments greater than 1 ha are never completely covered by buildings; therefore, a coverage value larger than 90% makes sure that built-up areas are excluded. Thirdly, a temporal difference of 7 years between the LiDAR data used for training (2014) and the PALSAR data (2007) used for application of the classification model exists. To make sure that a patch of woody vegetation identified by the CHM already existed in 2007, the thresholds, therefore, have to consider regrowth. The fastest growing vegetation type in New Zealand is pine tree plantations. Pinus radiata plantations typically grow 6 to 10 m in their first 7 years on the North Island.26 However, canopy closure would not typically reach 90% except in those stands planted with exceptionally high stocking density. Therefore, the 90% threshold makes sure the woody vegetation already existed at the time the PALSAR data were recorded. Histograms of per segment mean values of SPOT red, NIR and SWIR, NDVI, PALSAR HH and HV, and the HH/HV ratio were then created to examine the power of the seven predictors to discriminate between woody and nonwoody segments (Fig. 1). The histograms show that NDVI has very little ability to differentiate between woody and nonwoody vegetation, while all other predictors indicate some degree of discrimination between the two classes, with the best separation shown by SPOT SWIR and PALSAR HV. Fig. 1Histograms of woody (in green) versus nonwoody (in red) segments for each of the seven predictors. -axis for Satellite Pour l’Observation de la Terre (SPOT) red, near-infrared (NIR), and short-wave infrared (SWIR) bands shows reflectance values, phased array type L-band synthetic aperture radar (PALSAR) HH, and HV decibel (dB) of backscatter. -axis describes the number of segments. 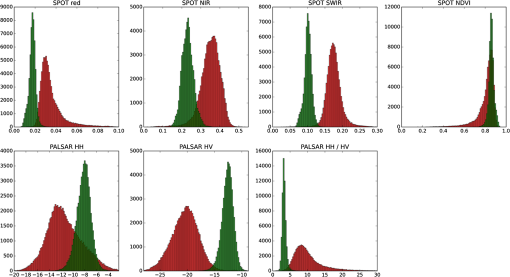 Scatterplots showing the two classes in a two-dimensional feature space were then created for all combinations of the seven predictors to visually check for potential correlation and none was detected. Figure 2 displays the three histograms showing the best differentiation based on SPOT only (red versus SWIR), PALSAR only (HH versus HV), and a combination of SPOT and PALSAR (SWIR versus HV). Fig. 2Scatterplots of woody (green) and nonwoody (red) segments for SPOT red versus SWIR band (reflectance), HH versus HV backscatter (dB), and SWIR band (reflectance) versus HV backscatter (dB). 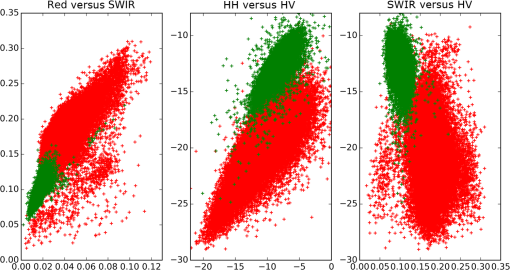 At this stage, we had identified six apparently uncorrelated predictors that had the ability to differentiate between woody and nonwoody segments with SPOT SWIR and PALSAR HV showing the best promise. We now had to find a suitable statistical model. Visual interpretation of scatterplots shows that a nonlinear differentiation function is preferable, which ruled out algorithms based on classification trees such as decision tress or random forest as well as discriminant analysis. Therefore, we decided to use SVM8 because of their versatility regarding the applied decision functions. Scikit-learn12 was chosen as the corresponding software library because of the familiarity of the authors with the python programming language and its well-structured architecture and documentation. To find the best SVM-based classifier, we compared model and cross-validation scores for different sets of predictors, different decision functions (linear, polynomial, and radial basis function), and parameterizations of the decision functions. Initial runs showed that linear as well as second- and third-degree polynomials performed best considering both physical interpretation of the discrimination function as well as model and cross-validation scores. The independent term of polynomial functions (coefficient 0) is by default set to 0 and we experimented with different values. Values of 0.05, 0.5, and 1 generally resulted in good results but performed differently for the various polynomial functions and predictor sets. Therefore, we evaluated all possible combinations of the mentioned values. The used predictor sets were as follows: 3.2.Classification of Identified Woody SegmentsTo include the identified woody segments into the next release of LCDB, it is necessary to classify them according to the existing scheme. The relevant classes were “Gorse and/or Broom,” “Mānuka and/or Kānuka,” “Broadleaved Indigenous Hardwoods,” “Mixed Exotic Shrubland,” “Matagouri or Gray Shrub,” “Deciduous Hardwoods,” “Indigenous Forest,” and “Exotic Forest.” We trained an SVM classifier based on the set of six predictors described earlier using segments with a majority coverage of the corresponding LCDB class. The classifier was then applied to the identified woody segments. 4.ResultsComparison of the different predictor sets and kernel functions showed that it is possible to develop a classifier using any of the four parameter sets. Highest scores and cross-validation scores () were achieved by the complete predictor set using third-degree polynomials and coefficient 0 set to 1.0. Scores of the SWIR/HV combination were only slightly lower. We decided to use the classifier with the overall highest score: the complete parameter set using a third-degree polynomial and a coefficient 0 of 1.0. For comparison and display purposes, we use the SWIR/HV predictor set with the same kernel parameters in parallel. To understand how the SVM classifier differentiates between the two target classes, we created a diagram showing the discrimination function in the two-dimensional case using the SWIR/HV predictor set (Fig. 3). The third-degree polynomial calculated by the SVM algorithm separates the two classes well. As shown in Fig. 3, the brown line is the discrimination function, if the woody and nonwoody classes use a weighting of 1:10. The reason for this weighting is that we want conservative identification of new woody patches, given that our general purpose is to improve the LCDB dataset. Therefore, introduction of segments that are incorrectly identified as woody is avoided. Weighting the two classes achieves this by moving the discrimination function closer to the woody cluster reducing the number or segments identified as woody and thus reducing false positives. Fig. 3Plot of nonweighted (yellow/blue area) and weighted (brown line) discrimination function between woody and nonwoody classes. -axis shows the SPOT SWIR band and -axis the PALSAR HV backscatter, both standardized (centered to 0 and scaled to unit variance). 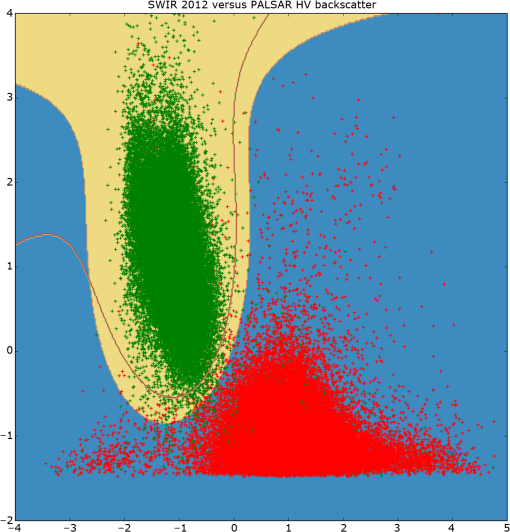 The woody/nonwoody classifier was trained with segments that were homogenous (defined by the 90% larger than the 3-m CHM-value threshold). To better understand the classifier when using heterogeneous segments, we applied it to all segments thus far classified by LCDB as pasture (LCDB classes “High-Producing Exotic Grassland,” “Low-Producing Grassland,” and “Depleted Grassland”), in the Wellington region where the LiDAR CHM was available. Overall, the Wellington region was divided into 272,856 segments. Of the segments previously identified by LCDB as pasture, 4733 segments were now identified as being woody by the classifier based on SWIR and HV backscatter only, while 4880 segments were identified using the classifier based on six predictors. Figure 4 shows the newly identified woody patches (in pasture) for the Wellington region, and Fig. 5 shows a subset of this. Fig. 4Land Cover Database (LCDB) map of the Wellington region, showing newly identified woody patches in pasture, as red. 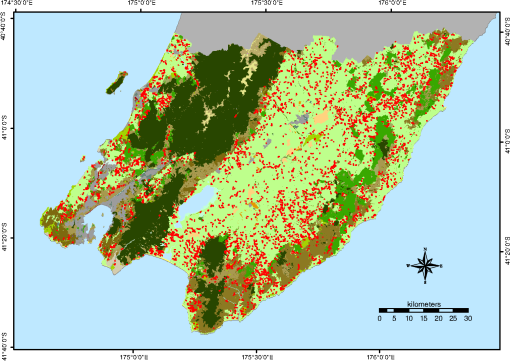 Fig. 5A subset of the Wellington region (LCDB), showing newly identified woody patches outlined in red. The woody patches are classified as outlined in Sec. 3.2. 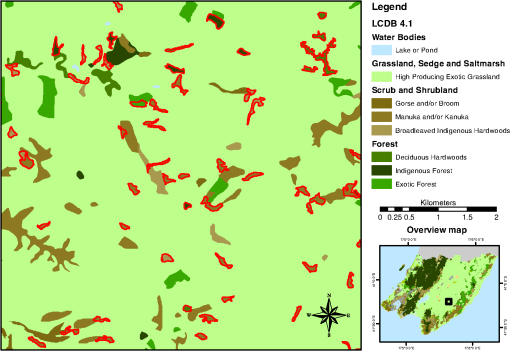 We then compared how many of the identified segments were also identified using CHM thresholds. For this, we identified those segments with a majority () covered by CHM values greater than 3, 2, 1, and 0.5 m, respectively (Table 1). According to this analysis, the classifier based on six predictors performed better than the one based on SWIR and HV when applied to real-world data, which is in accordance with model and cross-validation scores. For the height class, where the majority of woody vegetation has a height greater than 0.5 m, over 92% of the segments were correctly classified as woody. Table 1Comparison of number of segments identified by the SVC classifier with those identified by the light detection and ranging canopy height model (LiDAR CHM).
Both steps of the classifier were then applied to all 16 regions of New Zealand. The resulting raster clumps were polygonized and smoothed. To simplify inclusion in LCDB 4.1, the polygons were then filtered using a WITHIN spatial operation with existing pasture polygons. The output of this filter operation only included isolated and completely new polygons that could easily be checked by an operator before inclusion. Table 2 shows the number of these found in each region before and after filtering. The 16,840 isolated and smoothed polygons were visually checked against a temporal sequence of older Landsat7 imagery, the SPOT-5 imagery used in this study, more recent Landsat8 imagery, and more recent high-resolution SPOTMaps imagery (1.5 m). Each of these isolated woody features was assessed to see if it would be an improvement to the LCDB if included without any additional digitizing. The proportion considered useful was 76% (12,743). Those woody features considered to be an improvement were manually classified into LCDB categories by interpretation of the temporal image sequence and directly inserted into the LCDB. The prediction of which woody class the feature fell in was less accurate, in all 4627 (7783 ha) were correctly predicted or 36%. Table 2Statistics for discovered woody features with potential to improve LCDB. For each region, the first figure is the total number of woody features and the second figure is the reduction after filtering to ensure the feature is isolated and completely new to LCDB4.1.
5.DiscussionLand cover classification based on a combination of optical and SAR data depends on the capability of the input bands to discriminate between the target classes and a discrimination function that is able to take advantage of this discriminative power. Our results show that it is possible to create a successful classifier based on SPOT-5 or PALSAR dual-polarized data alone, but best results are achieved when the two data sources were combined. Model and cross-validation scores were very high () in the initial classification with two output classes. In the second classification step, achieved scores were much lower. This reflects the difficulty of differentiating between the multiple target classes and the quality of the LCDB used to train the classifier. The SVM algorithm used in this study was able to differentiate between the target classes with better results in the woody/nonwoody only case and shows general potential for analysis of combined datasets because of its flexibility regarding the applied discrimination function. Furthermore, it was helpful to be able to visualize the discrimination function, although only for two-dimensional parameter sets. This allowed compensating for one of the disadvantages of many machine-learning methods—their tendency to create black box models—and enabled us to understand and interpret the model from a physical perspective. Although we have shown that it is possible to add new information content to the existing LCDB dataset with synergetic usage of LiDAR, SPOT, and PALSAR data, manual examination of the output is still a necessity and will be in the future. We see potential, though, in refining and extending the defined methodology for future updates of LCDB. Manual work could then be redirected from on-screen drawing to quality assurance of automatically created improvement datasets. The developed method is applicable to other mapping projects on regional to national scales if the examined landscape is characterized by a mosaic of woody and nonwoody vegetation types. The approach used can also be applied to similar problems, e.g., the identification of clearings in woody areas. A more challenging problem that would need adjustment of the methodology would be to find segments of incorrectly identified vegetation where the discrimination is not clear-cut woody versus nonwoody. An example would be to identify forest areas on bush/scrub backgrounds. The training method of the SVM classifier using the CHM would have to be refined in this case and the temporal difference between the PALSAR and SPOT data would pose a greater problem than in this analysis. Current and temporally matching datasets for PALSAR, SPOT, and LiDAR CHM would generally be beneficial. The ALOS satellite that produced the PALSAR data used in this study ceased to function in 2011, but fortunately ALOS-2 started regular provision of PALSAR-227 data on December 1, 2014, with improved spatial resolution. Therefore, it should be possible to repeat the analysis described here in the future with more current SAR data. SPOT-6, on the other hand, no longer provides a SWIR band,28 which was found to be the most useful of the optical bands for woody/nonwoody discrimination. Therefore, the combination of PALSAR-2 and SPOT-6 might be a powerful combination for future vegetation classification, with PALSAR-2 compensating for the missing SWIR band. 6.ConclusionIt is possible to use a LiDAR CHM of a region as training data for a classifier. The classifier is an SVM applied to nationally available PALSAR and SPOT-5 imagery. Application of the classifier to segments increased the accuracy to levels suitable for use in enhancing the national LCDB (). Regional-scale differentiation between woody and nonwoody vegetation was best achieved by a combination of L-band dual-polarized PALSAR data (HV) with multispectral optical data that include an SWIR band. The application of an SVM algorithm to these data proved successful. The versatility of these algorithms regarding the discrimination function and their ability to solve classification problems with multiple output classes were critical factors for success. The identified and classified woody patches constitute a valuable addition and enhancement of the national LCDB. References, “LCDB v4.0—Land Cover Database version 4.0,”
(2014) https://lris.scinfo.org.nz/layer/412-lcdb-v40-land-cover-database-version-40/ May 2015). Google Scholar
G. Laurin et al.,
“Discrimination of vegetation types in alpine sites with ALOS PALSAR-, RADARSAT-2-, and LiDAR-derived information,”
Int. J. Remote Sens., 34 6898
–6913
(2013). http://dx.doi.org/10.1080/01431161.2013.810823 IJSEDK 0143-1161 Google Scholar
S. Erasmi and A. Twele,
“Regional land cover mapping in the humid tropics using combined optical and SAR satellite data: a case study from Central Sulawesi, Indonesia,”
Int. J. Remote Sens., 30
(10), 2465
–2478
(2009). http://dx.doi.org/10.1080/01431160802552728 IJSEDK 0143-1161 Google Scholar
H. T. Chu and L. L. Ge,
“Synergistic use of multi-temporal ALOS/PALSAR with SPOT multispectral satellite imagery for land cover mapping in the Ho Chi Minh city area, Vietnam,”
in IEEE Int. Symp. on Geoscience and Remote Sensing IGARSS,
1465
–1468
(2010). Google Scholar
L. Carvalho et al.,
“Optical and SAR imagery for mapping vegetation gradients in Brazilian savannas: synergy between pixel-based and object-based approaches,”
in Int. Conf. on Geographic Object-based Image Analysis,
1
–7
(2010). Google Scholar
E. Lehmann et al.,
“Joint processing of Landsat and ALOS-PALSAR data for forest mapping and monitoring,”
IEEE Trans. Geosci. Remote Sens., 50 55
–67
(2012). http://dx.doi.org/10.1109/TGRS.2011.2171495 Google Scholar
E. Lehman et al.,
“SAR and optical remote sensing: assessment of complementarity and interoperability in the context of a large-scale operational forest monitoring system,”
Remote Sens. Environ., 156 335
–348
(2015). http://dx.doi.org/10.1016/j.rse.2014.09.034 Google Scholar
C. Cortes and V. Vapnik,
“Support-vector networks,”
Mach. Learn., 20 273
–297
(1995). http://dx.doi.org/10.1007/BF00994018 Google Scholar
F. Melgani and L. Bruzzone,
“Classification of hyperspectral remote sensing images with support vector machines,”
IEEE Trans. Geosci. Remote Sens., 42
(8), 1778
–1790
(2004). http://dx.doi.org/10.1109/TGRS.2004.831865 Google Scholar
G. Camps-Valls et al.,
“Robust support vector method for hyperspectral data classification and knowledge discovery,”
IEEE Trans. Geosci. Remote Sens., 42
(7), 1530
–1542
(2004). http://dx.doi.org/10.1109/TGRS.2004.827262 Google Scholar
Y. Shao and R. Lunetta,
“Comparison of support vector machine, neural network and CART algorithms for the land-cover classification using limited training data points,”
ISPRS J. Photogramm. Remote Sens., 70 78
–87
(2012). http://dx.doi.org/10.1016/j.isprsjprs.2012.04.001 IRSEE9 0924-2716 Google Scholar
F. Pedregosa et al.,
“Scikit-learn: machine learning in python,”
J. Mach. Learn. Res., 12 2825
–2830
(2011). Google Scholar
G. Mountrakis, J. Im and C. Ogole,
“Support vector machines in remote sensing: a review,”
ISPRS J. Photogramm. Remote Sens., 66
(3), 247
–259
(2011). http://dx.doi.org/10.1016/j.isprsjprs.2010.11.001 IRSEE9 0924-2716 Google Scholar
T. Blaschke,
“Object based image analysis for remote sensing,”
ISPRS J. Photogramm. Remote Sens., 65 2
–16
(2010). http://dx.doi.org/10.1016/j.isprsjprs.2009.06.004 IRSEE9 0924-2716 Google Scholar
C. Gibbes et al.,
“Application classification and high resolution satellite imagery for Savanna ecosystem analysis,”
Remote Sens., 2 2748
–2772
(2010). http://dx.doi.org/10.3390/rs2122748 Google Scholar
R. M. Lucas et al.,
“Updating the Phase 1 habitat map of Wales, UK, using satellite sensor data,”
ISPRS J. Photogramm. Remote Sens., 66 81
–102
(2011). http://dx.doi.org/10.1016/j.isprsjprs.2010.09.004 IRSEE9 0924-2716 Google Scholar
B. W. Heumann,
“An object-based classification of Mangroves using a tree—support vector machine approach,”
Remote Sens., 3
(11), 2440
–2460
(2011). http://dx.doi.org/10.3390/rs3112440 Google Scholar
Indonesian National Standardisation Organisation (INSO), Kelas penutupan lahan dalam penafsiran citra optis resolusi sedan, Standar Nasional Indonesia, Jakarta, Indonesia
(2012). Google Scholar
J. D. Shepherd and J. R. Dymond,
“Correcting satellite imagery for the variance of reflectance and illumination of topography,”
Int. J. Remote Sens., 24 3503
–3514
(2003). http://dx.doi.org/10.1080/01431160210154029 IJSEDK 0143-1161 Google Scholar
M. Jakubauskas, K. Kindscher and D. Debinski,
“Spectral and biophysical relationships of montane sagebrush communities in multi-temporal SPOT XS data,”
Int. J. Remote Sens., 22
(9), 1767
–1778
(2001). http://dx.doi.org/10.1080/014311601300176042 IJSEDK 0143-1161 Google Scholar
M. Shimada and T. Ohtaki,
“Generating large-scale high-quality SAR mosaic datasets: application to PALSAR data for global monitoring,”
IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens., 3
(4), 637
–656
(2010). http://dx.doi.org/10.1109/JSTARS.2010.2077619 Google Scholar
M. Shimada et al.,
“PALSAR radiometric and geometric calibration,”
IEEE Trans. Geosci. Remote Sens., 47 3915
–3932
(2009). http://dx.doi.org/10.1109/TGRS.2009.2023909 Google Scholar
P. Bunting et al.,
“Sorted pulse data (SPD) library—Part II: a processing framework for LiDAR data from pulsed laser systems in terrestrial environments,”
Comput. Geosci., 7
(56), 207
–215
(2013). http://dx.doi.org/10.1016/j.cageo.2013.01.010 Google Scholar
D. Clewley et al.,
“A python-based open source system for geographic object-based image analysis (GEOBIA) utilizing raster attribute tables,”
Remote Sens., 6
(7), 6111
–6135
(2014). http://dx.doi.org/10.3390/rs6076111 Google Scholar
J. MacQueen,
“Some methods for classification and analysis of multivariate observations,”
in Proc. of the 5th Berkeley Symp. on Mathematical Statistics and Probability,
281
–297
(1967). Google Scholar
P. Peri et al.,
“Early growth and quality of radiata pine in a silvopastoral system in New Zealand,”
Agroforestry Syst., 55 207
–219
(2002). http://dx.doi.org/10.1023/A:1020588702923 Google Scholar
M. Shimada,
“ALOS-2 science program,”
in IEEE Int. Geoscience and Remote Sensing Symp. (IGARSS),
(2013). Google Scholar
, “SPOT-6 Satellite Sensor,”
(2015) http://www.satimagingcorp.com/satellite-sensors/spot-6/ October ). 2015). Google Scholar
BiographyMarkus U. Müller is an environmental informatics specialist on the informatics team, Landcare Research. He received his Dr. rer. nat. in geography from the University of Bonn in 2002. His research interests include remote sensing, geocomputation, and data science. James D. Shepherd is a scientist on the informatics team, Landcare Research. He received his PhD in remote sensing from the University of Waikato in 1997. He has researched methods for correcting satellite imagery for atmospheric and directional reflectance effects. He has been instrumental in developing cost-effective methods for mapping land cover and land use from satellite imagery in New Zealand, which are now used routinely in the LUCAS and Land Cover Database (LCDB) projects. He is currently the leader of the research stream of LCDB. John R. Dymond is a senior scientist on the soils and landscape team, Landcare Research. He received his DSc degree from the University of Canterbury in 2004 for the development of remote sensing for environmental monitoring. He is currently the leader of the Ecosystem Services for Multiple Outcomes program. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

